Evolutionary
Mathematical
Biology
Intragenomic conflict, genetics, epigenetics, epidemiology, evolutionary medicine, recombination and more...
About us
We, as individuals, are a collective of genes with each gene aiming to maximize its own evolutionary success.
As a result, the expression of genes can be detrimental for other genes, expressed in the same individual,
and may result in intragenomic conflict. This conflict can rip apart the individual causing among other things
health problems. We are interested in all kinds of intragenomic conflict and the detrimental effect these may
have in the individual. In particular, we are interested in detrimental effects with medical implications.
Our research focuses on genes that can kill other genes (meiotic drive), genes that can change other genes into
copies of themselves (gene conversion), genes that can move around the genome (transposable elements), genes that
can move across genomes (endogenous retroviruses) and genes that can keep memories of past hosts (imprinted genes).
We apply the gene’s eye view to understand maladaptive processes with medical implications, including infectious
diseases, recombination hotspots, genomic imprinting, cancer and sexual reproduction.
To explore this range of problems we use mathematical models of different types -including population genetics, kin selection, adaptive dynamics and game theory. We contrast the predictions of our mathematical models against large datasets.
Our Research
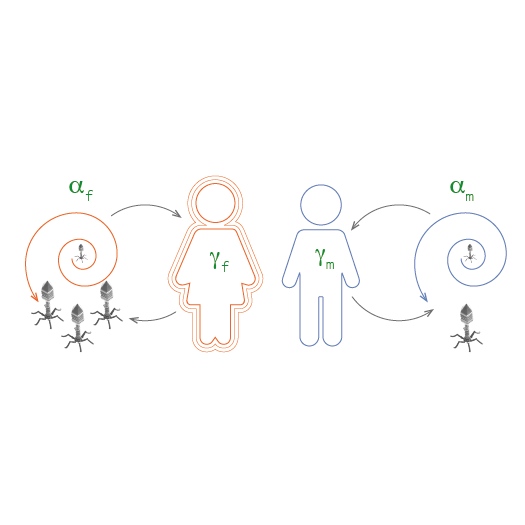
Differences between women and men in infectious diseases
Pathogens colonize hosts evolving and co-evolving with their genomes. We are particularly interested in researching how fundamental differences in the biology of females and males may lead to sex-specific differences in immune systems and the outcome of infectious diseases. From a broad perspective we are interested in contributing to developing a more effective treatment of infectious diseases in each of the sexes and addressing the difference in the incidence of autoimmune diseases and cancer in the sexes.
Recombination Hotspots
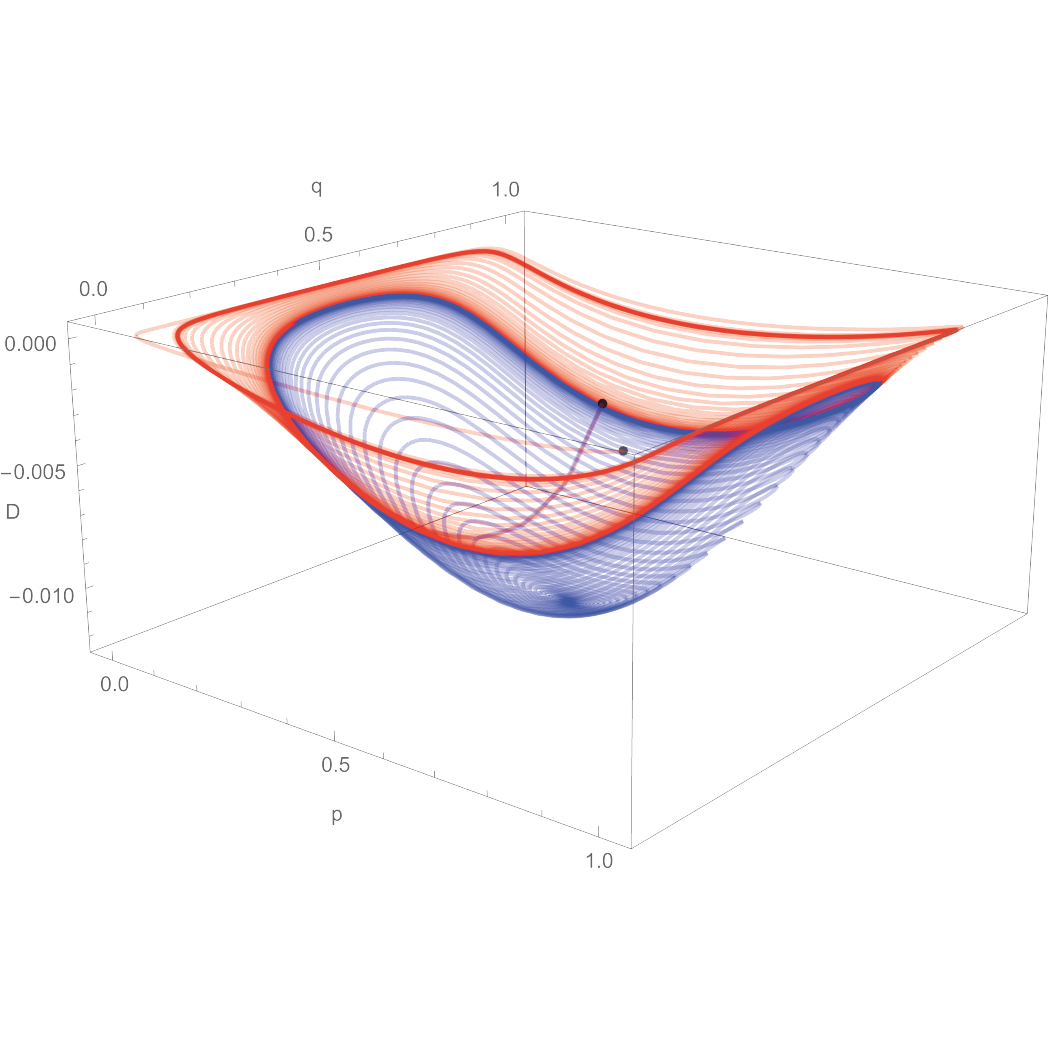
Recombination hotspots are regions where recombination is several orders of magnitude higher than the average in the genome. As part of the recombination process, the sequence that allows recombination is converted into the sequence that prevents recombination. This means that, over evolutionary time, recombination hotspots should be doomed to extinction, but they are not. We are interested in what evolutionary forces make possible the existence and ubiquity of recombination hotspots in mammals.
Menopause
Menopause is the transition from reproductive to non-reproductive life well before natural death. Rather than involving a smooth, rapid change, it is normally preceded by a long period of erratic hormonal fluctuation that is accompanied by a plethora of unpleasant symptoms, known as perimenopause. We are interested in the epigenetic mechanisms underpinning the menopausal transition and the variability of the menopause experience between different human populations.
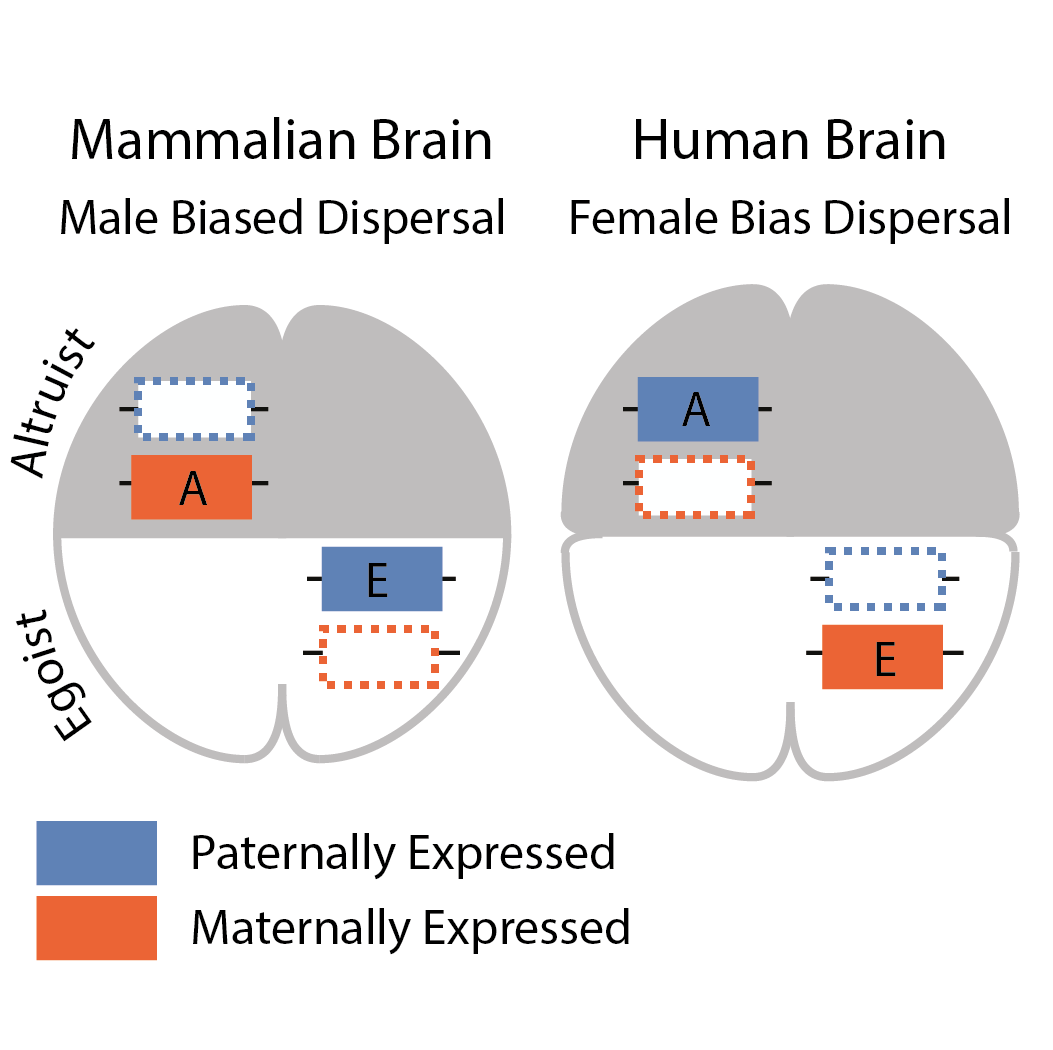
Genomic Imprinting
Imprinted genes are genes with different patterns of expression when inherited from females as opposed to males. They are found in humans and many other mammals. We are interested in exploring when does natural selection favour the evolution of genes that can remember which parent they came from.
Our collaborators include:
Our Members

Francisco Úbeda de Torres
I am a Reader at the School of Biological Sciences,
Royal Holloway, University of London.
My research focuses on developing mathematical models
to investigate conflict between genes expressed in the
same individual (intra-genomic conflict) and their
implications on disease. I am compelled to ensure that
my theoretical research has far-reaching impact and
broad appeal by applying it to understanding: the
epigenetic architecture of imprinted genes, the
persistence of recombination hotspots in human
populations, the difference in virulence of infections
in the sexes and the evolution of sexual reproduction…
among other topics.
The joy that research brings to my life (as captured in the
picture provided) is only comparable to the happiness that
the sun bestows on the estival evenings of London.

Frédéric Fyon
I have been a PostDoctorate Research Assitant with
Francisco since September 2020. Specialized in the use
of multi-locus population genetics models to
investigate evolutionary conundrums, I am interested in
everything that relates to reproduction and natural selection.
This includes topics as varied as the evolution of sex chromosomes,
of recombination hotspots, of asexual reproduction (with a special
focus in hybrid species), of hybrid genomes inheriting mutation
load from parental species with varied mating systems.
I did my phD at the
University of Montpellier under
the supervision of
Dr. Thomas Lenormand. Together, we developped a
theory of an unadaptative, indirect selective force
shaping the evolution of cis-regulators. This theory
notably led to an alternative explanation for the
divergence of sex chromosome .
In my research, I predominantly
use mathematical analysis and programming (C++, SLiM) of numerical
simulations.
To know more about me, here is my professional website.
Our Papers
-
On maternity and the stronger immune response in women.
E. Mitchell, A.L. Graham, F. Úbeda and G. Wild (2022) Nature Communications12:4273 -
Epigenetic Inheritance and the Evolution of Infectious Diseases
D. McLeod, G. Wild and F. Úbeda (2021) Nature Communications12:4273 -
Cycling under Quasi-Linkage Equilibrium
T. Russell. M. Russell, F. Úbeda and V. Jansen (2019) Journal of Theoretical Biology477:84-97 -
PRDM9 and the Evolution of Recombination Hotspots
F. Úbeda, T. Russell and V. Jansen (2019) Theoretical Population Biology126:19-32 -
The Meaning of Intragenomic Conflict
A. Gardner and F. Úbeda (2017) Nature Ecology & Evolution1:1807-1815 -
The Evolution of Sex-Specific Virulence in Infectious Diseases
F. Úbeda and V. Jansen (2016) Nature Communications7:13849 -
The Evolving Landscape of Imprinted Genes in Humans and Mice: Conflict among Alleles, Genes, Tissues and Kin
J. F. Wilkins, F. Úbeda and J. van Cleve (2016) BioEssays38:482-489 -
Intragenomic Conflict Over Dispersal
E. Farrell, F. Úbeda and A. Gardner (2015) American Naturalist186:E61-E71 -
Mother and Offspring in Conflict: Why Not?
F. Úbeda and A. Gardner (2015) PLoS Biology13:e1002084 -
On the Origin of Sex Chromosomes from Meiotic Drive
F. Úbeda, M. M. Patten & G. Wild (2015) Proc. Roy. Soc. B. 282:20141932 -
Specialists and Generalists: the Sexual Ecology of the Genome
D. Haig, F. Úbeda and M. M. Patten (2014) Cold Spring Harbor Perspectives 6:a017525 -
First Principles of Hamiltonian Medicine
B. Crespi, K. Foster and F. Úbeda (2014) Phil. Trans. R. Soc. B..369:20130366 -
Ecology Drives Intragenomic Conflict over Menopause
F. Úbeda, H. Ohtsuki and A. Gardner (2014) Ecology Letters17:165-174 -
Sexual and Parental Antagonism Shape Genomic Architecture
M. M. Patten, F. Úbeda and D. Haig (2013) Proc. Roy Soc. B. 280:20131795 -
A Model for Genomic Imprinting in the Social Brain: Elders
F. Úbeda and A. Gardner (2012) Evolution66:1567-1581 -
The Evolution of Sex
J. Engelstädter and F. Úbeda (2012). In: Encyclopedia of Theoretical Ecology. California University Press, Berkeley -
The Kinship Theory of Genomic Imprinting Evolving
Y. Brandvain, J. van Cleve, F. Úbeda and J.F. Wilkins (2011) Trends in Genetics27:251 -
Diseases Associated with Genomic Imprinting
J.F. Wilkins and F. Úbeda (2011) Progress in Molecular Biology and Translational Science -
Genomic Imprinting: an Obsession with Depilatory Mice
D. Haig and F. Úbeda (2011) Current Biology 21:R257-R259 -
Inclusive Fitness Theory and Eusocialy
P. Abbot, J. Abe, L. Alcock, S. Alizon et al. (2011) Nature 471:E1-E4 -
The Red Queen Theory of Recombination Hotspots
F. Úbeda and J.F. Wilkins (2011) Journal of Evolutionary Biology24:541-553 -
Power and Corruption
F. Úbeda and E.A. Duénez (2011) Evolution 65:1127-1139 -
A Model for Genomic Imprinting in the Social Brain: Adults
F. Úbeda and A. Gardner (2011) Evolution65:462-475 -
Genetic variation and community change: selection, evolution, and feedbacks
M.A. Genung, J.A. Schweitzer, F. Úbeda et al. (2011) Functional Ecology25:408-419 -
Let the Right One in: a Microeconomic Approach to Partner Choice
M. Archetti, F. Úbeda, D. Fudenberg et al (2011) American Naturalist 177: 75-85 -
Stable Linkage Disequilibrium Owing to Sexual Antagonism
F. Úbeda, D. Haig and M.M. Patten (2010) Proc. R. Soc. Lond. B278:855-862 -
Fitness Variation Due to Sexual Antagonism and Linkage Disequilibrium
M.M. Patten, D. Haig and F. Úbeda (2010) Evolution 64: 3638-3642 -
A Model for Genomic Imprinting in the Social Brain: Juveniles
F. Úbeda and A. Gardner (2010) Evolution 64: 2587-2600
Covered by the New York Times and Psychology Today -
From Genes to Ecosystems: Emerging Concepts Bridging Ecological and Evolutionary Dynamics
J.K. Bailey, J.A. Schweitzer, F. Úbeda et al (2010) In: The Ecology of Plant Secondary Metabolites: from Genes to Landscapes. Cambridge University Press, Cambridge. -
From Genes to Ecosystems: a Synthesis of the Effects of Plant Genetic Factors across Levels of Organization
J.K. Bailey, J.A. Schweitzer, F. Úbeda et al (2009) Phil. Trans. R. Soc. B. 364: 1607-1616 -
Evolution of Genomic Imprinting with Bi-parental Care: Implications for Prader-Willi and Angelman Syndromes
F. Úbeda (2008) PLoS Biology 6: 1678-1692
Recommended by F1000 and covered by the European Journal of Human Genetics . -
Imprinted Genes and Human Disease, an Evolutionary Perspective
F. Úbeda and J.F. Wilkins (2008) In: Genomic Imprinting. New York, Springer: 101-113 -
Male Killers and the Origins of Paternal Genome Elimination
F. Úbeda and B.B. Normark (2006) Theoretical Population Biology 70: 511-526 -
Why Mendelian Segregation?
F. Úbeda (2006) Biochemical Society Transactions 34: 566-568 -
On the Evolutionary Stability of Mendelian Segregation
F. Úbeda and D. Haig (2005) Genetics 170: 1345-1357 -
Sex-specific Meiotic Drive and Selection at an Imprinted Locus
F. Úbeda and D. Haig (2004) Genetics 167: 2083-95 -
Dividing the Child
F. Úbeda and D. Haig (2003) Genetica 117: 103-10 -
Clone Mixtures and a Pacemaker: New Faces of the Red-Queen Theory and Ecology
A. Sasaki, W.D. Hamilton and F. Úbeda (2002) Proc. R. Soc. Lond. B 269: 761-72
Contact Us
Disclaimer: your personal information as well as the content of this message will never be shared with any third party.